Tutorial¶
Getting started¶
Import library and packages¶
[1]:
import os
import sys
root_folder = os.path.dirname(os.getcwd())
sys.path.append(root_folder)
import ResoFit
from ResoFit.calibration import Calibration
from ResoFit.fitresonance import FitResonance
from ResoFit.experiment import Experiment
from ResoFit._utilities import Layer
import numpy as np
import matplotlib.pyplot as plt
Global paramters¶
min energy of 7 eV (has to be greater than 1 x 10-5 eV)
max energy of 150 eV (has to be less than 3000 eV)
energy steps to interpolate database: 0.1 eV
[2]:
energy_min = 7
energy_max = 150
energy_step = 0.01
File locations¶
data/* (directory to locate the file)
data_file (YOUR_DATA_FILE.txt or .csv)
spectra_file (YOUR_SPECTRA_FILE.txt or .csv)
[3]:
folder = 'data/_data_for_tutorial'
data_file = 'raw_data.csv'
spectra_file = 'spectra.txt'
Preview data using Experiment()¶
Note this is meant to demenstrate the functionality of Experiment() class. These operations are not needed for general calibration and fitting.
Data¶
Values are in transmission with image number as indexes after extracting with ImageJ
[5]:
experiment.data
[5]:
| 0 | |
|---|---|
| 0 | 1.025803 |
| 1 | 1.027222 |
| 2 | 1.029864 |
| 3 | 1.029095 |
| 4 | 1.032496 |
| 5 | 1.038683 |
| 6 | 1.042255 |
| 7 | 1.041075 |
| 8 | 1.034811 |
| 9 | 1.031851 |
| 10 | 1.032940 |
| 11 | 1.032937 |
| 12 | 1.034382 |
| 13 | 1.034205 |
| 14 | 1.035444 |
| 15 | 1.033757 |
| 16 | 1.039690 |
| 17 | 1.043947 |
| 18 | 1.043075 |
| 19 | 1.039344 |
| 20 | 1.039744 |
| 21 | 1.041730 |
| 22 | 1.041170 |
| 23 | 1.042075 |
| 24 | 1.040100 |
| 25 | 1.039565 |
| 26 | 1.039670 |
| 27 | 1.041171 |
| 28 | 1.044951 |
| 29 | 1.044430 |
| ... | ... |
| 2776 | 0.915787 |
| 2777 | 0.920761 |
| 2778 | 0.921717 |
| 2779 | 0.930883 |
| 2780 | 0.923302 |
| 2781 | 0.923815 |
| 2782 | 0.924068 |
| 2783 | 0.916594 |
| 2784 | 0.914615 |
| 2785 | 0.919048 |
| 2786 | 0.924325 |
| 2787 | 0.923643 |
| 2788 | 0.920561 |
| 2789 | 0.920570 |
| 2790 | 0.923603 |
| 2791 | 0.919968 |
| 2792 | 0.916965 |
| 2793 | 0.926718 |
| 2794 | 0.925573 |
| 2795 | 0.919824 |
| 2796 | 0.920326 |
| 2797 | 0.920852 |
| 2798 | 0.928154 |
| 2799 | 0.918875 |
| 2800 | 0.919932 |
| 2801 | 0.917441 |
| 2802 | 0.915185 |
| 2803 | 0.919612 |
| 2804 | 0.921282 |
| 2805 | 0.905286 |
2806 rows × 1 columns
Spectra¶
Column 0 time values (seconds) with image number as indexes
Column 1 neutron counts
[6]:
experiment.spectra
[6]:
| 0 | 1 | |
|---|---|---|
| 0 | 9.600000e-07 | 22192427 |
| 1 | 1.120000e-06 | 20966591 |
| 2 | 1.280000e-06 | 19678909 |
| 3 | 1.440000e-06 | 19890967 |
| 4 | 1.600000e-06 | 20968632 |
| 5 | 1.760000e-06 | 21026731 |
| 6 | 1.920000e-06 | 20464927 |
| 7 | 2.080000e-06 | 21112328 |
| 8 | 2.240000e-06 | 22520473 |
| 9 | 2.400000e-06 | 22849349 |
| 10 | 2.560000e-06 | 22234183 |
| 11 | 2.720000e-06 | 21483270 |
| 12 | 2.880000e-06 | 20741476 |
| 13 | 3.040000e-06 | 20045837 |
| 14 | 3.200000e-06 | 19278237 |
| 15 | 3.360000e-06 | 18293220 |
| 16 | 3.520000e-06 | 17256754 |
| 17 | 3.680000e-06 | 16811295 |
| 18 | 3.840000e-06 | 17305194 |
| 19 | 4.000000e-06 | 18163422 |
| 20 | 4.160000e-06 | 18526230 |
| 21 | 4.320000e-06 | 18573412 |
| 22 | 4.480000e-06 | 18822753 |
| 23 | 4.640000e-06 | 18829949 |
| 24 | 4.800000e-06 | 18510897 |
| 25 | 4.960000e-06 | 18082372 |
| 26 | 5.120000e-06 | 17696805 |
| 27 | 5.280000e-06 | 17336846 |
| 28 | 5.440000e-06 | 16983927 |
| 29 | 5.600000e-06 | 16640908 |
| ... | ... | ... |
| 2776 | 4.451200e-04 | 2467945 |
| 2777 | 4.452800e-04 | 2466877 |
| 2778 | 4.454400e-04 | 2469613 |
| 2779 | 4.456000e-04 | 2468271 |
| 2780 | 4.457600e-04 | 2471169 |
| 2781 | 4.459200e-04 | 2471734 |
| 2782 | 4.460800e-04 | 2466328 |
| 2783 | 4.462400e-04 | 2469199 |
| 2784 | 4.464000e-04 | 2466776 |
| 2785 | 4.465600e-04 | 2466053 |
| 2786 | 4.467200e-04 | 2476433 |
| 2787 | 4.468800e-04 | 2474714 |
| 2788 | 4.470400e-04 | 2469559 |
| 2789 | 4.472000e-04 | 2474567 |
| 2790 | 4.473600e-04 | 2471115 |
| 2791 | 4.475200e-04 | 2472997 |
| 2792 | 4.476800e-04 | 2476153 |
| 2793 | 4.478400e-04 | 2478613 |
| 2794 | 4.480000e-04 | 2474287 |
| 2795 | 4.481600e-04 | 2475409 |
| 2796 | 4.483200e-04 | 2478018 |
| 2797 | 4.484800e-04 | 2481255 |
| 2798 | 4.486400e-04 | 2484636 |
| 2799 | 4.488000e-04 | 2493780 |
| 2800 | 4.489600e-04 | 2499320 |
| 2801 | 4.491200e-04 | 2499035 |
| 2802 | 4.492800e-04 | 2509962 |
| 2803 | 4.494400e-04 | 2502085 |
| 2804 | 4.496000e-04 | 2488273 |
| 2805 | 4.497600e-04 | 11391066 |
2806 rows × 2 columns
Remove unwanted data points¶
[7]:
experiment.slice(slice_start=3, slice_end=2801, reset_index=False)
[7]:
(3 0.000001
4 0.000002
5 0.000002
6 0.000002
7 0.000002
8 0.000002
9 0.000002
10 0.000003
11 0.000003
12 0.000003
13 0.000003
14 0.000003
15 0.000003
16 0.000004
17 0.000004
18 0.000004
19 0.000004
20 0.000004
21 0.000004
22 0.000004
23 0.000005
24 0.000005
25 0.000005
26 0.000005
27 0.000005
28 0.000005
29 0.000006
30 0.000006
31 0.000006
32 0.000006
...
2771 0.000444
2772 0.000444
2773 0.000445
2774 0.000445
2775 0.000445
2776 0.000445
2777 0.000445
2778 0.000445
2779 0.000446
2780 0.000446
2781 0.000446
2782 0.000446
2783 0.000446
2784 0.000446
2785 0.000447
2786 0.000447
2787 0.000447
2788 0.000447
2789 0.000447
2790 0.000447
2791 0.000448
2792 0.000448
2793 0.000448
2794 0.000448
2795 0.000448
2796 0.000448
2797 0.000448
2798 0.000449
2799 0.000449
2800 0.000449
Name: 0, Length: 2798, dtype: float64, 3 1.029095
4 1.032496
5 1.038683
6 1.042255
7 1.041075
8 1.034811
9 1.031851
10 1.032940
11 1.032937
12 1.034382
13 1.034205
14 1.035444
15 1.033757
16 1.039690
17 1.043947
18 1.043075
19 1.039344
20 1.039744
21 1.041730
22 1.041170
23 1.042075
24 1.040100
25 1.039565
26 1.039670
27 1.041171
28 1.044951
29 1.044430
30 1.044017
31 1.045077
32 1.042383
...
2771 0.937393
2772 0.927724
2773 0.921393
2774 0.931766
2775 0.930592
2776 0.915787
2777 0.920761
2778 0.921717
2779 0.930883
2780 0.923302
2781 0.923815
2782 0.924068
2783 0.916594
2784 0.914615
2785 0.919048
2786 0.924325
2787 0.923643
2788 0.920561
2789 0.920570
2790 0.923603
2791 0.919968
2792 0.916965
2793 0.926718
2794 0.925573
2795 0.919824
2796 0.920326
2797 0.920852
2798 0.928154
2799 0.918875
2800 0.919932
Name: 0, Length: 2798, dtype: float64)
Retrieve values for X-axis¶
In ‘neutron energy’¶
[8]:
experiment.x_raw(offset_us=0., source_to_detector_m=16)
[8]:
array([ 6.45188613e+05, 5.22602776e+05, 4.31903121e+05, ...,
6.64684718e+00, 6.64210874e+00, 6.63737536e+00])
In ‘neutron wavelength’¶
[9]:
experiment.x_raw(angstrom=True, offset_us=0., source_to_detector_m=16)
[9]:
array([ 0.00035604, 0.0003956 , 0.00043516, ..., 0.11092624,
0.1109658 , 0.11100536])
Retrieve values for Y-axis¶
In ‘neutron attenuation’¶
[10]:
experiment.y_raw()
[10]:
array([-0.029095, -0.032496, -0.038683, ..., 0.071846, 0.081125,
0.080068])
In ‘neutron transmission’¶
[11]:
experiment.y_raw(transmission=True)
[11]:
array([ 1.029095, 1.032496, 1.038683, ..., 0.928154, 0.918875,
0.919932])
In ‘neutron attenuation’ & remove baseline¶
[12]:
experiment.y_raw(baseline=True)
[12]:
array([ 0.0183733 , 0.01488139, 0.00860353, ..., 0.01160011,
0.02088986, 0.01984365])
Retrieve interpolated values for both X-axis & Y-axis¶
X-axis: ‘neutron energy’
Y-axis: ‘neutron attenuation’
[13]:
experiment.xy_scaled(energy_min=energy_min, energy_max=energy_max, energy_step=energy_step,
angstrom=False, transmission=False,
offset_us=0, source_to_detector_m=15,
baseline=True)
[13]:
(array([ 7. , 7.01, 7.02, ..., 149.98, 149.99, 150. ]),
array([ 0.00584673, -0.00275185, 0.00486153, ..., 0.01071007,
0.01063659, 0.01056193]))
Plot raw in various ways¶
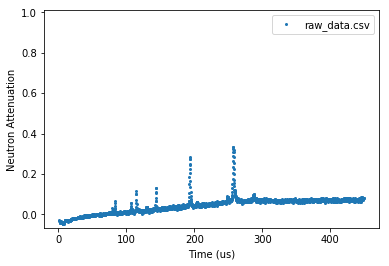
Attenuation vs. recorded time¶
Parameters need to be specified: - time_unit from ['s', 'us', 'ns']. (‘us’) is default
[15]:
experiment.plot_raw(x_axis='time')
plt.show()
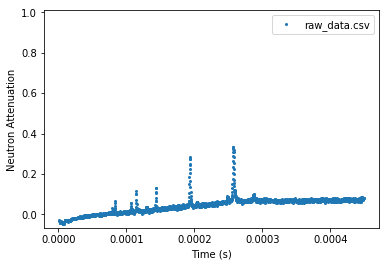
[16]:
experiment.plot_raw(x_axis='time', time_unit='s')
plt.show()
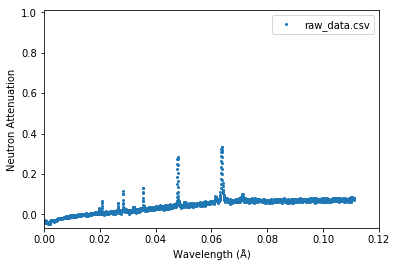
Attenuation vs. neutron wavelength¶
Parameters need to be specified: - lambda_xmax in (angstrom) - offset_us in (us) - source_to_detector_m in (m)
[17]:
experiment.plot_raw(x_axis='lambda', lambda_xmax=0.12,
offset_us=0, source_to_detector_m=16)
plt.show()
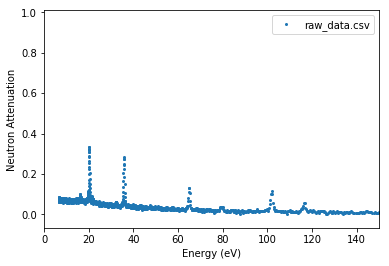
Attenuation vs. neutron energy¶
Parameters need to be specified: - energy_xmax in (eV) - offset_us in (us) - source_to_detector_m in (m)
[18]:
experiment.plot_raw(x_axis='energy', energy_xmax=150,
offset_us=0, source_to_detector_m=16)
plt.show()
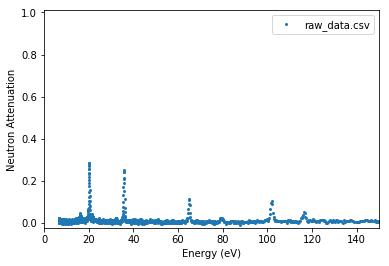
Remove baseline for plot¶
Note: this only changes the display of data, not the data values.
[19]:
experiment.plot_raw(offset_us=0, source_to_detector_m=16,
x_axis='energy', baseline=True,
energy_xmax=150)
plt.show()
Operation to experiment data¶
experiment.norm_to(file=background.csv)
This can be used to further normalize signal. (data divide background)
Note: this will change the data values.
Input sample info¶
Gd and U mixed foil
thickness neutron path within the sample in (mm)
density sample density in (g/cm3), if omitted, pure solid density will be used in fitting
repeat : reptition number if the data is summed of multiple runs (default: 1)
[20]:
layer_1 = 'U'
thickness_1 = 0.05 # mm
density_1 = None # g/cm^3 (if None or omitted, pure solid density will be used in fitting step)
layer_2 = 'Gd'
thickness_2 = 0.05 # mm
density_2 = None # g/cm^3 (if None or omitted, pure solid density will be used in fitting step)
Create sample layer
Note: Input each element as single layer.
[21]:
layer = Layer()
layer.add_layer(layer=layer_1, thickness_mm=thickness_1, density_gcm3=density_1)
layer.add_layer(layer=layer_2, thickness_mm=thickness_2, density_gcm3=density_2)
Calibration¶
Estimated intrumental parameters¶
input estimated source to detector distance (m)
input estimated possible time offset in spectra file (us)
Note: a separated Experiment() will be created after the initialization of Calibration(). Calibration.slice() is needed if you would like to remove unwanted data points by indexes.
[22]:
source_to_detector_m = 16.
offset_us = 0
Class initialization¶
Pass all the parameters definded above into the Calibration()
[23]:
calibration = Calibration(data_file=data_file,
spectra_file=spectra_file,
raw_layer=layer,
energy_min=energy_min,
energy_max=energy_max,
energy_step=energy_step,
folder=folder,
baseline=True)
Equations for (time-wavelength-energy) conversion¶
\(E\) : energy in (meV),
\(\lambda\) : wavelength in (Å).
\(t_{record}\) : recorded time in (µs),
\(t_{offset}\) : recorded time offset in (µs),
\(L\) : source to detector distance in (cm).
Calibrate instrumental parameters¶
using source_to_detector_m or offset_us or both to minimize the difference between the measured resonance signals and the simulated resonance signals from ImagingReso within the range specified in global parameters
vary can be one of [‘source_to_detector’, ‘offset’, ‘all’] (default is ‘all’)
fitting parameters are displayed
[24]:
calibration.calibrate(source_to_detector_m=source_to_detector_m,
offset_us=offset_us,
vary='all')
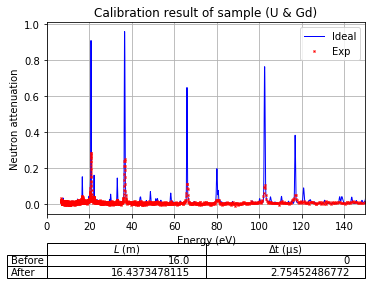
-------Calibration-------
Params before:
Name Value Min Max Stderr Vary Expr Brute_Step
offset_us 0 -inf inf None True None None
source_to_detector_m 16 -inf inf None True None None
Params after:
Name Value Min Max Stderr Vary Expr Brute_Step
offset_us 2.755 -inf inf 0.03047 True None None
source_to_detector_m 16.44 -inf inf 0.002896 True None None
Calibration chi^2 : 51.508119690122165
[24]:
<lmfit.minimizer.MinimizerResult at 0x119da62b0>
Retrieve calibrated parameters¶
[25]:
calibration.calibrated_offset_us
[25]:
2.754524867719895
[26]:
calibration.calibrated_source_to_detector_m
[26]:
16.437347811454231
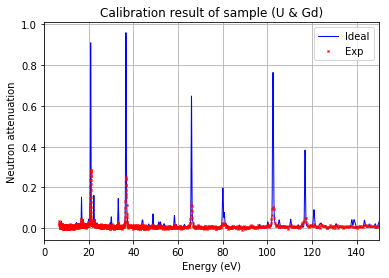
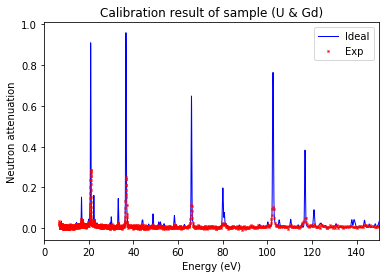
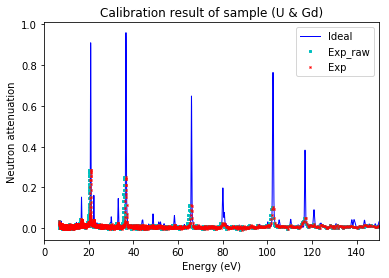
Plot calibration result¶
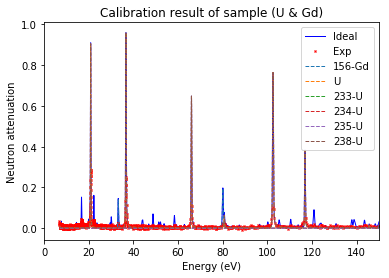
using the best fitted source_to_detector_m and offset_us to show the calibrated resonance signals from measured data and the expected resonance positions from ImagingReso
measured data before and after is ploted with raw data points instead of interpolated data points. However, the interpolated data was used during the calibration step above.
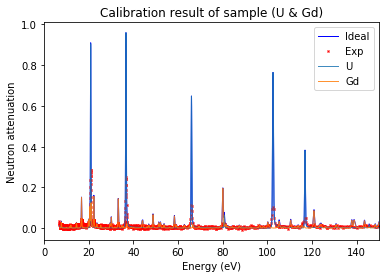
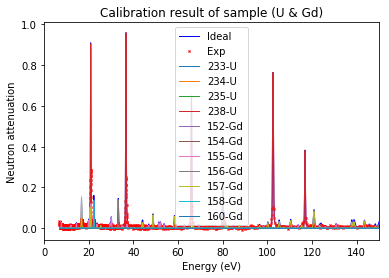
Show specified items¶
Format as list with each name in string
[33]:
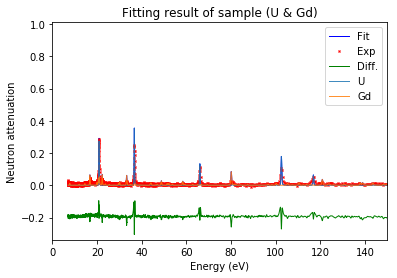
calibration.plot(table=False, grid=False, items_to_plot=['U', 'U-238', 'Gd-156'])
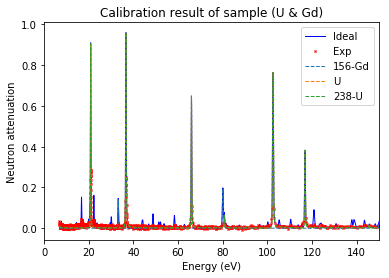
Note: 'U*' can be used to represent all the isotopes of uranium
[34]:
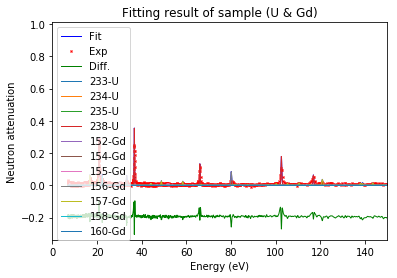
calibration.plot(table=False, grid=False, items_to_plot=['U', 'U*', 'Gd-156'])
Fit resonances¶
Class initialization¶
Pass all the parameters definded and calibrated into the FitResonance()
[35]:
fit = FitResonance(spectra_file=spectra_file,
data_file=data_file,
folder=folder,
energy_min=energy_min,
energy_max=energy_max,
energy_step=energy_step,
calibrated_offset_us=calibration.calibrated_offset_us,
calibrated_source_to_detector_m=calibration.calibrated_source_to_detector_m,
baseline=True)
Fitting equations¶
Beer-Lambert Law:¶
\({ N }_{ i }\) : number of atoms per unit volume of element \(i\),
\({ d }_{ i }\) : effective thickness along the neutron path of element \(i\),
\({ \sigma }_{ ij }\left( E \right)\) : energy-dependent neutron total cross-section for the isotope \(j\) of element \(i\),
\({ A }_{ ij }\) : abundance for the isotope \(j\) of element \(i\).
\({N_A}\) : Avogadro’s number,
\({C_i}\) : molar concentration of element \(i\),
\({\rho_i}\) : density of the element \(i\),
\(m_{ij}\) : atomic mass values for the isotope \(j\) of element \(i\).
How to fit the resonance signals¶
using thickness (mm) or density (g/cm3) to minimize the difference between the measured resonance signals and the simulated resonance signals from ImagingReso within the range specified in global parameters
vary can be one of [‘thickness’, ‘density’] (default is ‘density’)
fitting parameters are displayed
[36]:
fit_result = fit.fit(layer, vary='density')
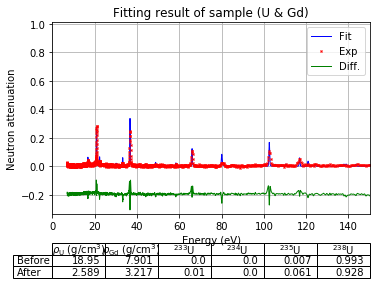
-------Fitting (density)-------
Params before:
Name Value Min Max Stderr Vary Expr Brute_Step
density_gcm3_Gd 7.901 0 inf None True None None
density_gcm3_U 18.95 0 inf None True None None
thickness_mm_Gd 0.05 0 inf None False None None
thickness_mm_U 0.05 0 inf None False None None
Params after:
Name Value Min Max Stderr Vary Expr Brute_Step
density_gcm3_Gd 3.217 0 inf 0.04812 True None None
density_gcm3_U 2.589 0 inf 0.01818 True None None
thickness_mm_Gd 0.05 0 inf 0 False None None
thickness_mm_U 0.05 0 inf 0 False None None
Fitting chi^2 : 2.7164917570446554
Output fitted result in molar concentration¶
unit: mol/cm3
[37]:
fit.molar_conc()
Molar-conc. (mol/cm3) Before_fit After_fit
U 0.07961217820137899 0.010874916048358519
Gd 0.05024483306836248 0.020460587861440557
[37]:
{'Gd': {'density': {'units': 'g/cm3', 'value': 3.2174274412115276},
'layer': 'Gd',
'molar_conc': {'units': 'mol/cm3', 'value': 0.020460587861440557},
'molar_mass': {'units': 'g/mol', 'value': 157.25},
'thickness': {'units': 'mm', 'value': 0.05}},
'U': {'density': {'units': 'g/cm3', 'value': 2.5885444133322855},
'layer': 'U',
'molar_conc': {'units': 'mol/cm3', 'value': 0.010874916048358519},
'molar_mass': {'units': 'g/mol', 'value': 238.02891},
'thickness': {'units': 'mm', 'value': 0.05}}}
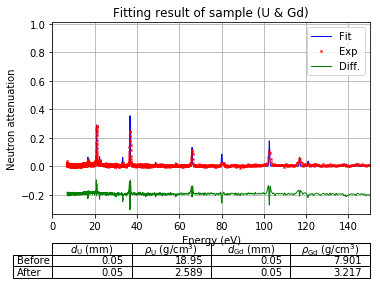
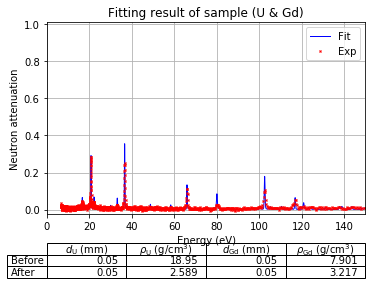
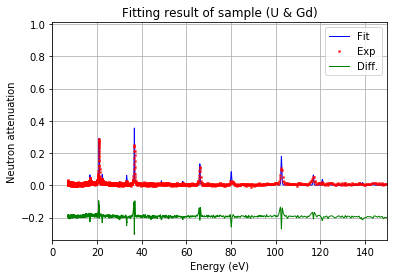
Plot fitting result¶
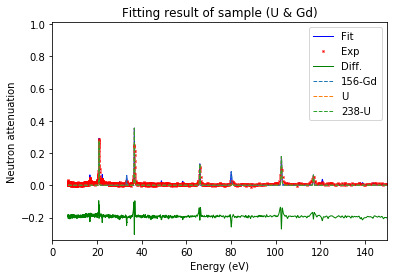
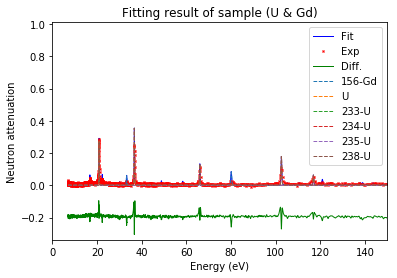
using the best fitted density to show the measured resonance signals and the fitted resonance signals from ImagingReso
measured data before and after is ploted with raw data points instead of interpolated data points. However, the interpolated data was used during the fitting step above.
Show specified items¶
Format as list with each name in string
[44]:
fit.plot(table=False, grid=False, items_to_plot=['U', 'U-238', 'Gd-156'])
[45]:
fit.plot(table=False, grid=False, items_to_plot=['U', 'U*', 'Gd-156'])
Fit isotopic ratios¶
specify the element you would like to perform isotopic (by at.%) fitting.
Note: Processing time of this step might be long.
[46]:
fit.fit_iso(layer='U')
-------Fitting (isotopic at.%)-------
Params before:
Name Value Min Max Stderr Vary Expr Brute_Step
U233 0 0 1 None True None None
U234 5.5e-05 0 1 None True None None
U235 0.0072 0 1 None True None None
U238 0.9927 0 1 None False 1-U233-U234-U235 None
Params after:
Name Value Min Max Stderr Vary Expr Brute_Step
U233 0.0101 0 1 0.03743 True None None
U234 5.316e-06 0 1 0.04623 True None None
U235 0.06146 0 1 0.03617 True None None
U238 0.9284 0 1 0.05946 False 1-U233-U234-U235 None
Fit iso chi^2 : 2.700599444474776
[47]:
fit.molar_conc()
Molar-conc. (mol/cm3) Before_fit After_fit
U 0.07968367366422123 0.01088468223204817
Gd 0.05024483306836248 0.020460587861440557
[47]:
{'Gd': {'density': {'units': 'g/cm3', 'value': 3.2174274412115276},
'layer': 'Gd',
'molar_conc': {'units': 'mol/cm3', 'value': 0.020460587861440557},
'molar_mass': {'units': 'g/mol', 'value': 157.25},
'thickness': {'units': 'mm', 'value': 0.05}},
'U': {'density': {'units': 'g/cm3', 'value': 2.5885444133322855},
'layer': 'U',
'molar_conc': {'units': 'mol/cm3', 'value': 0.01088468223204817},
'molar_mass': {'units': 'g/mol', 'value': 237.81534069141117},
'thickness': {'units': 'mm', 'value': 0.05}}}
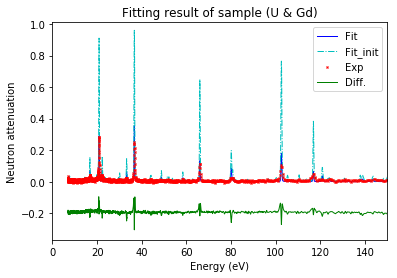
[48]:
fit.plot()